» How is Cornelia de Lange Syndrome Diagnosed

Genetic Testing for the CdLS Spectrum (post birth)
Genetic tests have been developed to identify changes in any of the genes associated with the CdLS spectrum. A referral to a clinical geneticist can be made if parents feel that it would be useful to discuss the possibility of testing for the child with a CdLS spectrum syndrome. Genetic testing is not always appropriate or necessary (particularly if a doctor is very certain about the clinical diagnosis), but would be the only way to approach prenatal testing for future pregnancies.
Panel Sequencing
The most effective genetic test to identify a change in a gene known to cause CdLS is panel sequencing. Panel sequencing analyses multiple genes at once. When testing for CdLS spectrum the panel should include at least the seven known CdLS genes (see Figure 5 on page 16). Most laboratories for genetic testing will include additional genes that can cause a phenotype resembling CdLS, such as CREBBP and EP300.
Sanger Sequencing
Although panel sequencing is the most effective genetic test, it may not be available in some parts of the world. Clinical geneticists may use other genetic tests, such as Sanger sequencing. Sanger sequencing uses a computer to look for gene mutations in a person’s DNA. If an individual has a classic CdLS phenotype, Sanger sequencing of the NIPBL gene is the preferred initial test. If an individual has a non-classic CdLS phenotype, other genes associated with the CdLS spectrum can be sequenced (R9).
Testing for Mosaicism
When a change in a gene associated with CdLS cannot be found using panel or Sanger sequencing, genetic testing can evaluate an individual’s DNA for mosaicism. Mosaicism is where there are different populations of cells which have a different genetic make-up in a single person. See the Mosaicism section on page 12.
Multiplex Ligation-dependent Probe Amplification
If genetic testing is not able to find mosaicism in an individual’s DNA, a test called multiplex ligation-dependent probe amplification (MLPA) can be considered. MLPA looks for deletions or duplications in the NIPBL gene.
|
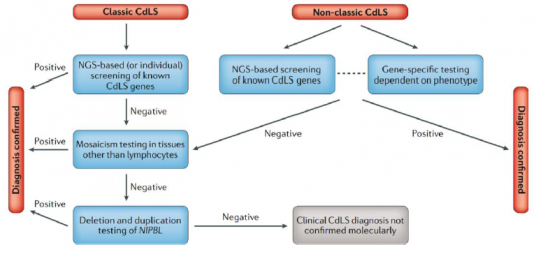
NGS = next generation sequencing/modern genetic testing approaches Fig. 5 | Diagnostic pathways for Cornelia de Lange (CdLS) Syndrome. In individuals with the classic phenotype, the first-line approach should be modern genetic testing approaches by either examining multiple genes at once, examining all of the genes that code for proteins in an individual or by examining the complete DNA of an individual. This should include all of the currently known CdLS genes (NIPBL, SMC1A, SMC3, RAD21, BRD4, HDAC8 and ANKRD11). If these approaches are not available, genetic testing should begin by examining NIPBL (classic CdLS phenotype). In individuals with the non-classic CdLS phenotype, the phenotype itself may allow experienced clinicians to determine which gene should be sequenced first; if this cannot be determined, modern genetic testing approaches can be performed. In the case of negative results, NIPBL and the other CdLS genes should be tested for mosaicism using tissues other than blood (as not all of the cells tested may show the mutation), for example, cells in connective tissues, cheek/mouth swabs or cells from urine. There are other tests that can be carried out to investigate the NIPBL gene (i.e. whether it has been deleted or duplicated), these are called multiplex ligation-dependent probe amplification or chromosome microarray. These tests can be used before other genetic testing approaches, or may be used following these approaches to investigate gene variants further. |
NGS = next generation sequencing/modern genetic testing approaches
Fig. 5 | Diagnostic pathways for Cornelia de Lange (CdLS) Syndrome.
In individuals with the classic phenotype, the first-line approach should be modern genetic testing approaches by either examining multiple genes at once, examining all of the genes that code for proteins in an individual or by examining the complete DNA of an individual. This should include all of the currently known CdLS genes (NIPBL, SMC1A, SMC3, RAD21, BRD4, HDAC8 and ANKRD11). If these approaches are not available, genetic testing should begin by examining NIPBL (classic CdLS phenotype). In individuals with the non-classic CdLS phenotype, the phenotype itself may allow experienced clinicians to determine which gene should be sequenced first; if this cannot be determined, modern genetic testing approaches can be performed. In the case of negative results, NIPBL and the other CdLS genes should be tested for mosaicism using tissues other than blood (as not all of the cells tested may show the mutation), for example, cells in connective tissues, cheek/mouth swabs or cells from urine. There are other tests that can be carried out to investigate the NIPBL gene (i.e. whether it has been deleted or duplicated), these are called multiplex ligation-dependent probe amplification or chromosome microarray. These tests can be used before other genetic testing approaches, or may be used following these approaches to investigate gene variants further.